CanGraph.GraphifyDrugBank package
In this page
CanGraph.GraphifyDrugBank package#
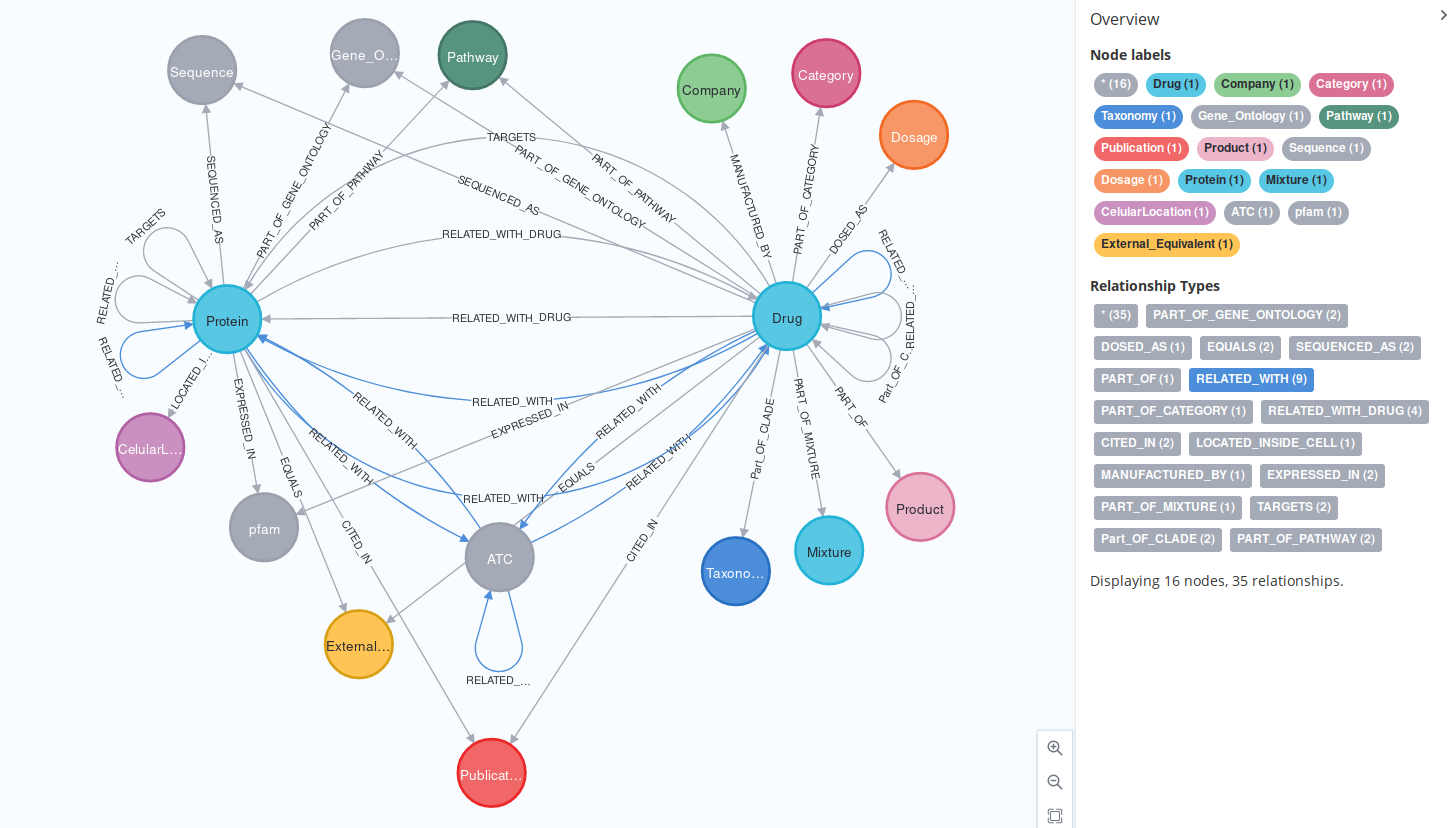
This package, created as part of my Master’s Intenship at IARC, imports nodes from the DrugBank Database (a high quality database containing a drugs and proteins with some characteristics) to Neo4J format in an automated way, providing an export in GraphML format.
To run, it uses alive_progress to generate an interactive progress bar (that shows the script is still running through its most time-consuming parts) and the neo4j python driver. This requirements can be installed using: pip install -r requirements.txt.
To run the script itself, use:
python3 main.py neo4jadress username databasepassword filename
where:
neo4jadress: is the URL of the database, in neo4j:// or bolt:// format
username: the username for your neo4j instance. Remember, the default is neo4j
password: the passowrd for your database. Since the arguments are passed by BaSH onto python3, you might need to escape special characters
filename: the database’s file name. For practical purpouses, it must be present in ./xmlfolder
Please note that there are two kinds of functions in the associated code: those that use python f-strings, which themselves contain text that cannot be directly copied into Neo4J (for instance, double brackets have to be turned into simple brackets) and normal multi-line strings, which can. This is because f-strings allow for variable customization, while normal strings dont.
An archived version of this repository that takes into account the gitignored files can be created using: git archive HEAD -o ${PWD##*/}.zip
Important Notices#
To access the Drugbank database, you have to previously request access at: https://go.drugbank.com/public_users/sign_up
Some XML tags have been intentionally not processed; for example, the
tag didn’t seem to dd anything new, and the and tags are likely not relevant for our project. The tag has no info (check cat full\ database.xml | grep "<ahfs-codes>") and thetag seemed like too much work (same for ). nOT USE because not of use to us Some :Proteins might also be :Drugs and viceversa: this makes the schema difficult to understand, but provides more meaningful data. This is also why some relations (
RELATED_WITH, for instance) dont have a relation: this way, we avoid duplicates.
The package consists of the following modules:
CanGraph.GraphifyDrugBank.build_database module#
A python module that provides the necessary functions to transition the DrugBank database to graph format,
either from scratch importing all the nodes (as showcased in CanGraph.GraphifyDrugBank.main) or in a case-by-case basis,
to annotate existing metabolites (as showcased in CanGraph.main).
- add_atc_codes(filename)[source]#
Creates “ATC” nodes based on XML files obtained from the DrugBank website. These represent the different ATC codes a Drug can be related with (including an small taxonomy)
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_categories(filename)[source]#
Creates “Category” nodes based on XML files obtained from the DrugBank website. These represent the different MeSH IDs a Drug can be related with
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Each category seems to have an associated MeSH ID. Maybe could rename nodes as MeSH?
- add_dosages(filename)[source]#
Creates “Dosage” nodes based on XML files obtained from the DrugBank website. These represent the different Dosages that a Drug should be administered at.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Warning
Using CREATE might generate duplicate nodes, but there was no unique characteristic to MERGE nodes into.
- add_drug_interactions(filename)[source]#
Creates
`(d)-[r:RELATED_WITH_DRUG]-(dd)`interactions between “Drug” nodes, whether they existed before or not. These are intentionally non-directional, as they should be related with each other.- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_drugs(filename)[source]#
Creates “Drug” nodes based on XML files obtained from the DrugBank website, adding some essential identifiers and external properties.
See also
This way of working has been taken from William Lyon’s Blog
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Since Publications dont have any standard identificator, they are created using the “Title”
- add_experimental_properties(filename)[source]#
Adds some experimental properties to existing “Drug” nodes based on XML files obtained from the DrugBank website.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_external_equivalents(filename)[source]#
Adds some external equivalents to existing “Drug” nodes based on XML files obtained from the DrugBank website. This should be “exact matches” of the Drug in other databases.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
The main reason to add them as “External-Equivalents” is because I felt these IDs where of not much use (and are thus easier to eliminate due to their common label)
- add_external_identifiers(filename)[source]#
Adds some external identifiers to existing “Drug” nodes based on XML files obtained from the DrugBank website.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
These also adds a “Protein” label to any “Drug”-labeled nodes which have a “UniProtKB”-ID among their properties. NOTE that this can look confusing in the DB Schema!!!
- add_general_references(filename)[source]#
Creates “Publication” nodes based on XML files obtained from the DrugBank website.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
Since not all nodes present a “PubMed_ID” field (which would be ideal to uniquely-identify Publications, as the “Text” field is way more prone to typos/errors), nodes will be created using the “Authors” field. This means some duplicates might exist, which should be accounted for.
- add_manufacturers(filename)[source]#
Creates “Company” nodes based on XML files obtained from the DrugBank website. These represent the different Companies that manufacture a Drug’s compound (not just package it)
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_mixtures(filename)[source]#
Creates “Mixture” nodes based on XML files obtained from the DrugBank website. These are the mixtures of existing Drugs, which may or may not be on the market.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
This doesn’t seem of much use, but has been added nonetheless just in case.
- add_packagers(filename)[source]#
Creates “Company” nodes based on XML files obtained from the DrugBank website. These represent the different Companies that package a Drug’s compounds (not the ones that manufacture them)
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
- add_pathways_and_relations(filename)[source]#
Adds “Pathway” nodes based on XML files obtained from the DrugBank website. It also adds some relations between Drugs and Proteins (which, remember, could even be the same kind of node) It is also able to tag both a Protein’s and a Drug¡s relation with a given Pathway In general, a Pathway involves a collection of Enzymes, Drugs and Proteins, with a SMPDB_ID (cool for interconnexion!)
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Warning
This function uses a “double UNWIND” clause, which means that we are only representing <pathways> tags with <enzymes> tags inside. Fortunately, this seems to seldom not happen, so it should represent no problem.
- add_products(filename)[source]#
Creates “Product” nodes based on XML files obtained from the DrugBank website. These are the individal medicaments that have been approved (or not) by the FDA
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Warning
Using CREATE means that duplicates will appear; unfortunately, I couldnt any unique_id field to use as ID when MERGEing the nodes. This should be accounted for.
- add_sequences(filename)[source]#
Creates “Sequence” nodes based on XML files obtained from the DrugBank website. These represent the AminoAcid sequence of Drugs that are of a peptidic nature.
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Todo
In some other parts of the script, sequences are being added as properties on Protein nodes. A common format should be set.
- add_targets_enzymes_carriers_and_transporters(filename, tag_name)[source]#
A REALLY HUGE function. It takes a filename and a tag_name, and gets info and creates “Protein” nodes with tag_name set as their role. It also adds a bunch of additional info, such as Publications, Targets, Actions, GO_IDs, PFAMs and/or some External IDs
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
tag_name (str) – The type of Protein node you want to import; it must be one of [“enzymes”, “carriers”, “transporters”] It is recommended that you run this function thrice, once for each type of protein
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Warning
We are using a bunch of concatenated UNWINDs, which force the existance of all elements in the UNWIND chain. This might remove some elements, but this is a HUUUUUGE database, and, to be honest, most things seem to almost always be present. An example is References and Polypeptides; Since there seem to be more References that Polypeptides, we try to UNWIND those first. The same can be said on the rest of UNWINDS: as we have external-id >>>>>>>>>> go-classifier >>>> pfam >> synonyms (in order of occurrence. not number of tags) , we UNWIND in that order to mitigate data loss
Note
To fix repetitions in properties such as Actions or Synonyms (caused by the HUGE number of UNWINDs), we tried lots of different strategies, finally coming up with SET p.Synonyms = replace(p.Synonyms, synonym._text + “,”, “”). This is cool! But means there will always be a trailing comma (removing it was not easy in this same transaction, though it could (TODO?) be done at the end.
Note
<tag_name>
- add_taxonomy(filename)[source]#
Creates “Taxonomy” nodes based on XML files obtained from the DrugBank website. These represent the “kind” of Drug we are dealing with (Family, etc)
- Parameters
tx (neo4j.Session) – The session under which the driver is running
filename (str) – The name of the XML file that is being imported
- Returns
A Neo4J connexion to the database that modifies it according to the CYPHER statement contained in the function.
- Return type
Note
It only creates relationships in the Kingdom -> Super Class -> Class -> Subclass direction, and from any node -> Drug. This means that, if any member of the Kingdom -> Super Class -> Class -> Subclass is absent, the line will be broken; hopefully in that case a new Drug will come in to rescue and settle the relation!
Warning
Some nodes without labels might be created if names are null: This has to be accounted for later on in the process
- build_from_file(newfile, driver)[source]#
A function able to build a portion of the DrugBank database in graph format, provided that one XML is supplied to it. This can either be the
`full_database.xml`file that you can get in DrugBank’s website, or a splitted version of it, with just one item per file (which is recommended due to memory limitations)- Parameters
newfile (str) – The path of the XML file to import
driver (neo4j.Driver) – Neo4J’s Bolt Driver currently in use
- Returns
This function modifies the Neo4J Database as desired, but does not produce any particular return.
CanGraph.GraphifyDrugBank.main module#
A python module that leverages the functions present in the build_database
module to recreate the DrugBank database using a graph format
and Neo4J, and then provides an GraphML export file.
Please note that, to work, the functions here pre-suppose you have a DrugBank account with access to the whole Database.
You have to prevously apply for it at: https://go.drugbank.com/public_users/sign_up, and place the `full_database.xml`
file at `./xmlfolder`.
For more details on how to run this script, please consult the package’s README
